A computational model has been created to simulate the biomechanical growth of breast tumors
Research by UC3M and Johns Hopkins University
3/20/24
Scientists from Universidad Carlos III de Madrid (UC3M) and Johns Hopkins University (JHU), in the USA, have analyzed the growth of breast tumors from a biomechanical perspective and have created a computational model that simulates the invasion process of cancer cells, depending on the characteristics of the surrounding tissue and cell junctions, among other parameters. This type of model will help predict the evolution of a tumor in patients from its mechanical properties (stiffness, density, etc.) of the surrounding microenvironment, which can be determined through a biopsy or imaging techniques.
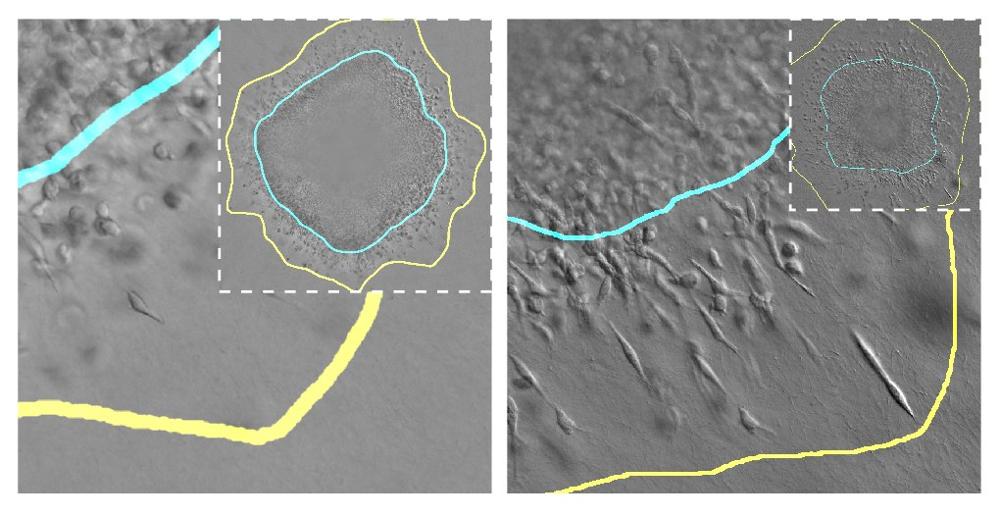
Tumor cells invading surrounding tissue under different mechanical conditions. The blue line indicates the boundary of the solid tumor and the yellow line indicates the space invaded by cells that detach from the tumor.
The growth process of a solid tumor involves its expansion through the surrounding tissue, usually composed of a fibrillar matrix (for example, collagen). Its expansion depends on many factors such as the total number of tumor cells, their volume and stiffness, their access to nutrients, and the mechanical properties of the tissue in which they are developing. Supported by experimental in vitro models, these UC3M and JHU researchers have developed a model that allows for simulating the evolution of the tumor growth on a computer, taking these factors into account. “In this model we have simulated how breast tumor cells invade the surrounding tissue, and how they proliferate more or less depending on how stiff and porous the surrounding tissue is or how strong the cell junctions with other cells are”, explains one of the researchers, Daniel García González, Associate Professor in UC3M's Continuum Mechanics and Structural Analysis Department and head of the ERC 4D-BIOMAP project.
To do this, the researchers have worked with spheroids to simulate how cells behave in a real tumor under different mechanical conditions. These spheroids consist of groups of tumor cells embedded in a fibrillar matrix whose characteristics can be modulated. “They are very powerful systems that are increasingly being used to study tumor behavior and to study possible therapies”, explains another of the researchers, Arrate Muñoz-Barrutia, a Professor in UC3M's Bioengineering Department.
Thanks to these spheroids, researchers have been able to modify certain biological or mechanical aspects of these tumors in the laboratory and evaluate how these variables influence cell proliferation and migration. They then transformed these observations into mathematical equations implemented in a computational model. In this way, they were able to test in parallel (in the computer simulator and in the experimental model with the spheroids in the laboratory) the variables that influence the growth of these tumors. “Our new multi-compartment spheroid system allowed us to control and modulate the system’s biomechanical properties via collagen density and E-cadherin expression, which are known to play a role in breast cancer progression. It was very exciting to work with this team to see the story come together from both experimental and computational perspectives”, says another of the study's authors, Denis Wirtz, from JHU's Chemical and Biomolecular Engineering Department.
“While experimentally, proliferation and invasion are often measured as two independent parameters, we observed a strong coupling of these processes. Although they could not be isolated using traditional experimental outputs, the computational model allowed us to study these processes independently and gather insights from the biomechanical properties of our system”, adds another of the JHU team's researchers, Ashleigh Crawford.
Future applications of this study are promising, according to the researchers. “If we know which mechanical parameters determine whether the tumor grows more or less, then we could use that data to improve treatment or develop new drugs in the medium or long term”, says Daniel García González. “We think that these studies open the door to the development of technologies that allow us to characterize the mechanics of the tumor, which can add relevant information for the choice of cancer therapy”, adds Arrate Muñoz-Barrutia.
The team of scientists also highlights the importance of multidisciplinary research in this case, since contributions have been made from computational and mathematical to purely biological fields. “My training as a biomedical engineer, studying at UC3M, has allowed me to collaborate in all parts of this research and to create bridges of communication between disciplines that use different terminologies”, says another of the study's authors, Clara Gómez Cruz, a PhD student in UC3M's Continuum Mechanics and Structural Analysis Department.
This research is part of 4D-BIOMAP (Biomechanical Stimulation based on 4D Printed Magneto-Active Polymer), a project funded by the European Research Council through an ERC Starting Grant from the European Union's Framework Programme for Research and Innovation, Horizon 2020 (GA 947723). It has also received funding from the USA's National Institute of Health and National Cancer Institute.
Bibliographic reference:
Crawford A.J. Gomez-Cruz, C. Russo G. C. Huang, W. Bhorkar I. Roy, T, Muñoz-Barrutia, A. Wirtz, D. Garcia-Gonzalez, D. (2024). Tumor proliferation and invasion are intrinsically coupled and unraveled through tunable spheroid and physics-based models. Acta Biomaterialia, Volume 175, Pages 170-185, ISSN 1742-7061. https://doi.org/10.1016/j.actbio.2023.12.043